HitGen has established a drug discovery research platform for small molecules and nucleic acid drugs centered on the design, synthesis and screening of DNA encoded chemical libraries (DELs), fragment-based drug discovery (FBDD) and structure-based drug design (SBDD) technologies.
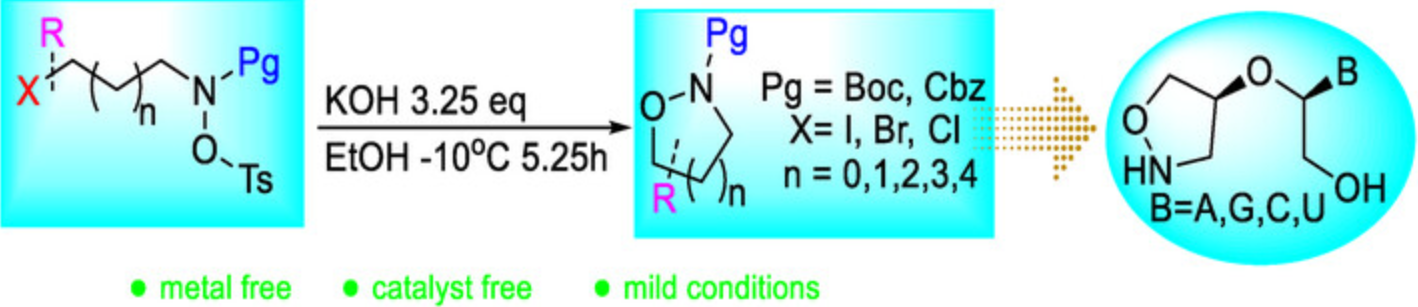
N–O heterocycles are privileged scaffolds in medicinal chemistry and materials science, yet their synthesis often requires harsh conditions or metal catalysts, limiting functional group compatibility. Herein, we report a mild, metal-free intramolecular cyclization strategy for the efficient assembly of diverse N–O heterocycles (4- to 8-membered rings). A sequential optimization approach employing definitive screening, steepest ascent, and full factorial designs significantly enhanced the reaction yield up to 95%. The method exhibits broad substrate scope, tolerating halogens, alkyl/aryl groups, and sensitive functionalities, and enables the synthesis of complex fused and nucleic acid-derived heterocycles. Its utility is demonstrated through the efficient preparation of unprotected unlocked nucleic acid (UNA) analogs, underscoring potential for therapeutic oligonucleotide development.
A metal-free intramolecular cyclization under mild conditions enables efficient synthesis of diverse N–O heterocycles (4–8 membered rings). Optimized via DoE, the method offers broad functional group tolerance and high yields, and is successfully applied in the concise preparation of bioactive unlocked nucleic acid (UNA) analogs.Graphical Abstract
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information