Get up to date with the latest HitGen articles and join us in the events
As one of the largest and most functionally diverse gene families, kinases have been recognized as key regulators/mediators of biological signaling networks. More than 30% of drug development efforts target these enzymes in the past two decades. As of January 2024, more than 90 small molecule kinase drugs have been approved by the US Food and Drug Administration (FDA), with approximately 200 orally available protein kinase inhibitors in clinical trials. These compounds have had a significant impact on the treatment of both oncological and non-oncological conditions.
Among the small molecule kinase drugs approved by the FDA, more than 50 are classified as multi-target drugs, representing the majority on the market (Figure 1 A). While the multi-target approach offers a broader therapeutic spectrum and enhances treatment flexibility and efficiency by acting on multiple related pathways, it also presents potential drawbacks. These may include increased risks of off-target effects and toxicity due to the broader activity profile, which can complicate the management of side effects and patient care. Only less than 40 of the FDA approved drugs are single-target kinase drugs, focusing on crucial kinase targets such as EGFR (Epidermal Growth Factor Receptor), BTK (Bruton's Tyrosine Kinase), BCR-ABL (Breakpoint Cluster Region-Abelson). These drugs demonstrate precision in targeting specific disease pathways, offering targeted therapies with potentially fewer side effects due to their specific mechanism of action. According to Cortellis CCI, 15 kinase drugs achieved sales exceeding 1 billion dollars in 2023, including 2 BCR-ABL inhibitors, 3 CDK4/6 (Cyclin-dependent Kinase 4/6) inhibitors, 2 BTK inhibitors, 3 JAK (Janus Kinase) inhibitors, and 5 other inhibitors targeting EGFR, BRAF (Rapidly Accelerated Fibrosarcoma B-type), ALK (Anaplastic Lymphoma Kinase), MET (Mesenchymal to Epithelial Transition factor) and VEGFR (Vascular Endothelial Growth Factor Receptor), respectively (Figure 1 B). The top-selling drugs, Ibrutinib, Palbociclib, and Ruxolitinib, are all first-in-class medications. Moreover, Alectinib, approved in 2015, has become the best-in-class drug for treating ALK-positive non-small cell lung cancer (NSCLC); Abemaciclib, approved in 2017, is believed to have the potential to surpass Palbociclib as the best-in-class treatment for ER-/HER2- (Estrogen Receptor-/Human Epidermal growth factor Receptor 2-)breast cancer.
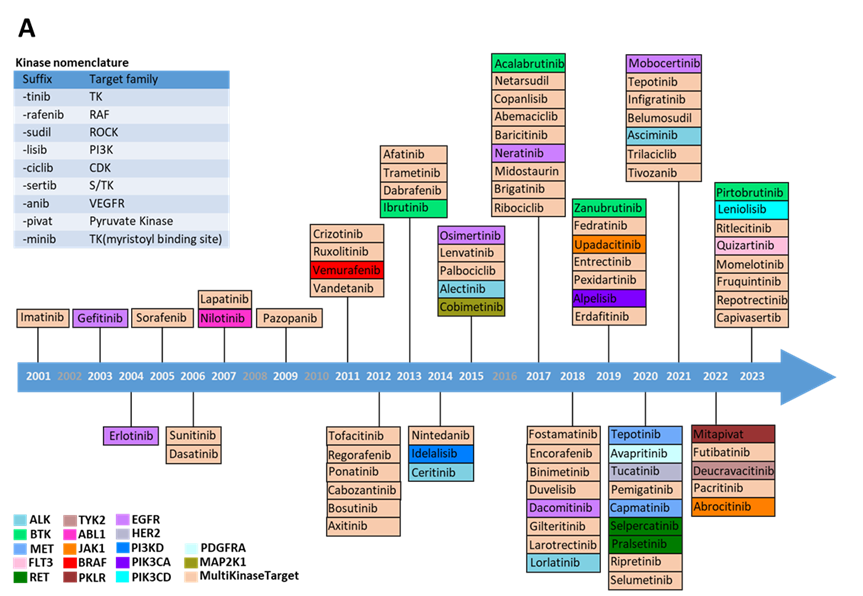
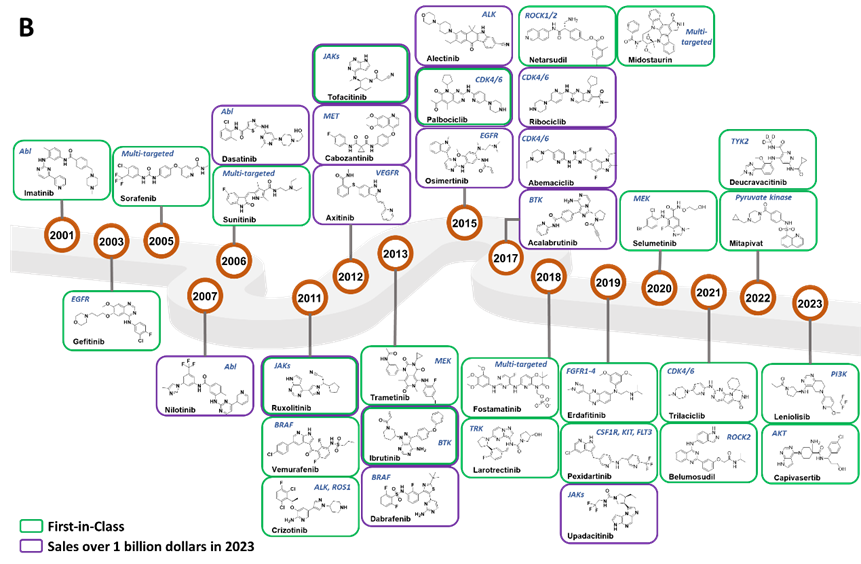
Figure 1. FDA approved small molecule kinase drugs since 2001. A) Drugs categorized by target types. Drugs are ordered by the year of approval and drug targets are represented by boxes with different colors. B) Structures of representative drugs. The first-in-class drugs are in green boxes and drugs with sales over $1 billion in 2023 are in purple boxes, the main targets are labeled.
Key challenges for kinase inhibitor discovery
Selectivity
Due to the high degree of homology in the ATP-binding sites, achieving selectivity and circumventing off-target interactions in kinase inhibitor design has been a significant challenge. The simultaneous inhibition of several protein kinases may bring potential advantages by targeting more than one target, however, this is more often related to toxicity and unwanted side effects. For example, the first-generation JAK inhibitors (Tofacitinib, Incyte's Ruxolitinib, Baricitinib, Peficitinib, and Delgocitinib) are pan-JAK inhibitors, exhibiting similar interaction patterns with JAK1, JAK2, JAK3, and TYK2. They bind within a hydrophobic pocket composed of residues L881, A906, V938, L959, and L1010 (based on JAK1 numbering) and form hydrogen bonds with hinge region residues (Figure 2 A). These non-selective inhibitors target multiple JAKs and block several associated signaling pathways, which leads to severe side effects including infections and anemia. However, through continuous research and development, several second-generation JAK inhibitors have been approved up to date (Figure 2 B), they selectively inhibit JAK family members, thereby suppressing specific disease-related signaling pathways while preserving the function of other cytokines.

Figure 2. A) Tofacitinib forms two hydrogen bonds with hinge region residues of JAK1 (PDB: 3EYG). B) Chemical structures of representative JAK Small Molecule Drugs.
Resistance
With the emergence of kinase mutants, the kinase inhibitors become less efficacious and newer generation inhibitors are needed. Next-generation sequencing is utilized to detect the driver mutations and structural analysis of the mutant sheds light on potential drug design strategies. In the case of EGFR, first and second generation inhibitors were approved for the treatment of non-small cell lung cancer. However, the T790M mutation, with increased affinity for ATP, exhibited resistance and made the inhibitors ineffective, the inefficacy of the first and second generation inhibitors necessitated the development of the third generation covalent inhibitors, specifically targeting the 797 Cysteine (Figure 3). Nevertheless, the emergence of new resistance mutant C797S abolished the covalent formation. Currently, there is a pressing need for fourth-generation EGFR inhibitors that can overcome the C797S mutation, which is still in the developmental stage. Therefore, the requirement for effective hit-finding tools for the discovery and development of kinase inhibitors against the mutants, meanwhile keeping good selectivity over kinase panel, is paramount.
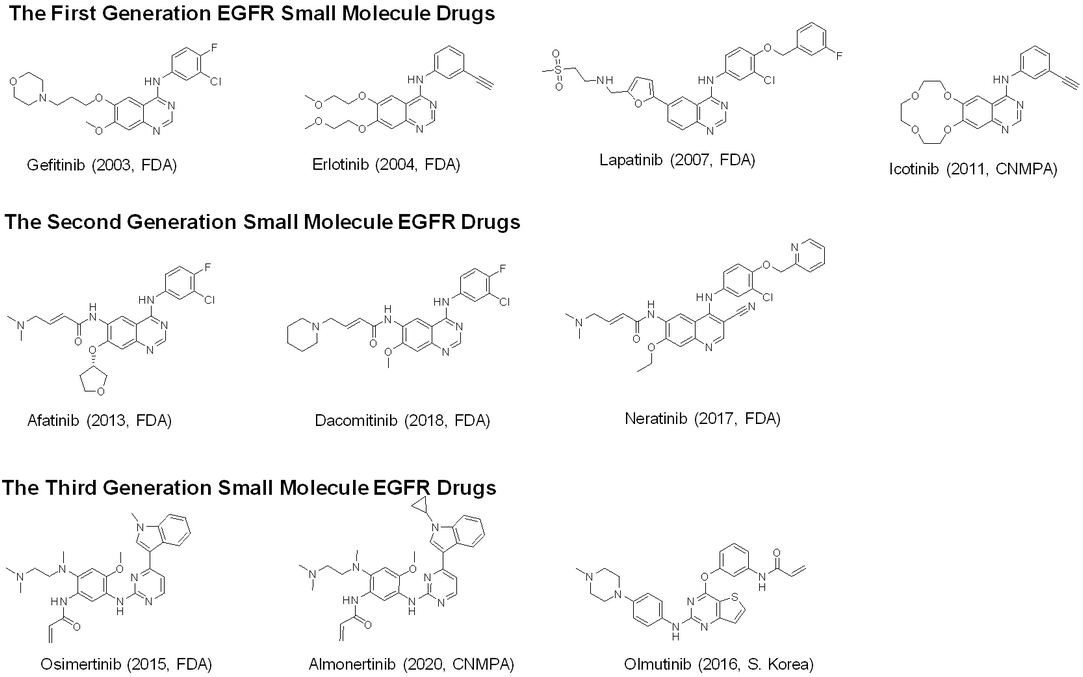
Figure 3. Chemical structures of representative EGFR Small Molecule Drugs.
DNA encoded library (DEL) technology, as a structural hypothesis free approach, is capable of screening billion to trillion diverse molecules and potentially identify compounds selectively interacting with the target kinase. This is exemplified by the discovery of the Receptor Interacting Protein 1 (RIP1) Kinase inhibitor, with minor optimization of the original hit from DEL, the candidate GSK2982772 progressed into clinical trial (J. Med. Chem. 2017, 60, 4, 1247–1261) (Figure 4). Actually, kinase inhibitor discovery via DEL screening has gained a lot of traction recently, which is evidenced by many HitGen partners’ kinase inhibitor discovery projects. Our comprehensive understanding of kinase biology and rich screening data provide a very effective way of identifying potent and selective inhibitors for more kinases.
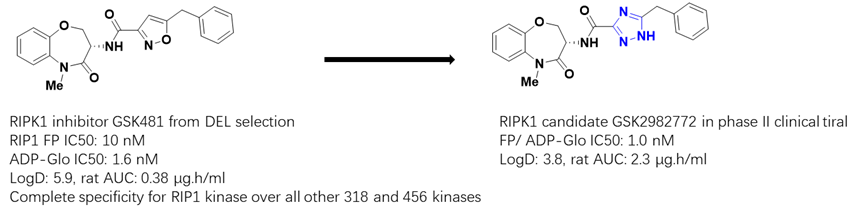
Figure 4. RIPK1 inhibitor discovered from DEL selection and the optimization to clinical candidate.
“Smart Selection Campaign” for inhibitor selectivity
As one of the key advantages for DEL technology, multiple selection experiments could be conducted in parallel. With our “Smart Selection Campaign” strategy, the selectivity of DEL compounds against different kinases can be defined at the very beginning. For example, a tyrosine kinase target has multiple family siblings who share a highly conserved carboxyl-terminal kinase domain and a relatively unique amino-terminal domain. In order to achieve good selectivity of the compounds against this particular kinase, four closely related kinases (A-D) were included in the DEL selection campaign as counter-screen samples. Upon filtering out all overlapped binders, specific binders can be selected for synthesis and validation. The selection results of these 5 samples and compound validation confirming good potency and selectivity are shown in Figure 5.
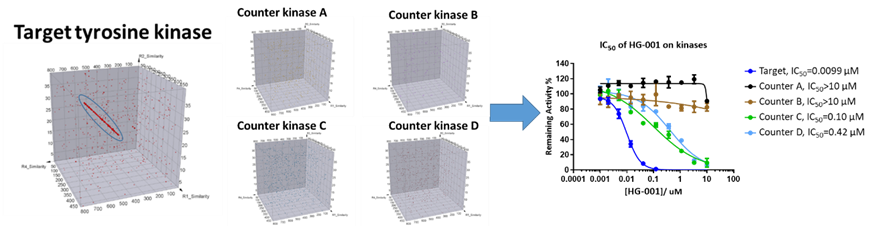
Figure 5. DEL selection results including target kinase as well as four count-screen kinase targets and compound validation for potency and selectivity.
Identification of kinase inhibitors with different MoA and mutant selectivity
As a mature target class of drug discovery, kinase inhibitors for the treatment of diverse types of diseases have proven successful. The most recent effort has also been extended to exploration of unique MoA (Mechanisms of Action) with kinase inhibitors. Tool compound(s) properly conjugating with DNA tag(s) could be used as spike-in reference(s) in the DEL selection to assess the pocket accessibility. With potent tool compound(s) occupying pocket(s) of interest, we are able to fine tune the pocket/binding site mapping for DEL selection (Figure 6A). By incorporating the mutants and wild type proteins in the same selection campaign and using the known selective inhibitor to differentiate potential binding sites, mutant selective kinase inhibitors were identified (Figure 6B).
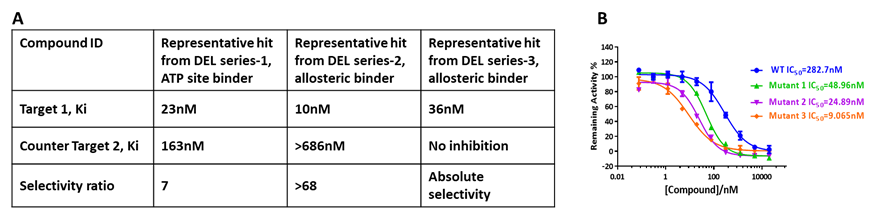
Figure 6. Multiple hits with different MoA (A) and mutant selectivity (B) have been identified against different kinases.
Targeting different kinase subfamilies with catalytic domain and/or regulatory domain
Kinases can be further classified according to their capability to phosphorylate different residues of the substrates, the kinases screened with HitGen DELs are summarized in Figure 7A. Regardless of their subtypes, almost all kinases share a similar catalytic domain and many kinases also contain regulatory domains. Therefore, a fundamental question to discover a kinase inhibitor is whether to target the catalytic domain or the regulatory (non-kinase) region.
Lipid kinases: The intrinsic structural differences of lipid kinases with protein kinases provide possibilities to identify selective inhibitors. The regulatory subunits of lipid kinases usually harbor binding pockets for potential small molecule inhibitors. This offers a good opportunity to identify novel kinase inhibitors, not only because of their pocket accessibility, but also the potential better selectivity by avoiding targeting the more conserved catalytic domains.
Pseudokinases: Pseudokinases usually harbor similar 3D structures as their catalytically active siblings, making them potentially attractive targets for DEL selection. Indeed, occupancy by competitive ligands of the ATP binding sites could result in potential interruption of the downstream pathway, whereas the conformation changes introduced by the allosteric ligands would likely lead to regulatory disruption, and these binders could be further developed into targeted degraders. By focusing on the non-catalytic pseudokinase domain, HitGen has identified multiple series of hits that have been later confirmed to be direct regulatory interrupters (Figure 7B).
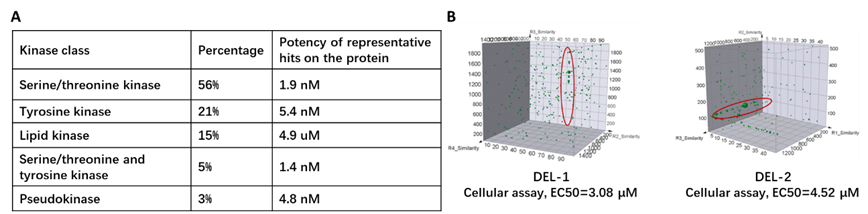
Figure 7. A) Distribution of kinase subtypes screened at HitGen and potency of representative hits. B) Pseudokinase domain-focused DEL selection for kinase subfamilies with hits identified.
Discover your kinase inhibitors at HitGen
Discovery of kinase inhibitors is about compound potency, selectivity, novelty and patentability. With our proprietary fine-tuned trillion libraries, rich DEL selection experiences, utilization of accumulated selection data as filters, the identification of kinase inhibitors using HitGen DELs has proven to be fast, cost effective and most importantly guarantees success rate.

We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information