One-stop Technology Platform to Accelerate Drug Discovery and Development
Enzymes are arguably the most ideal targets for small molecule drugs, owing to their involvement in chemical reaction catalysis and the better ligandability of the binding pockets. Although high-throughput screening (HTS) of inhibitors for enzyme targets has been well established, DNA encoded library (DEL) selection holds unique advantages, especially for those enzyme targets with multi-binding pockets. This is because the different conformations and binding pockets of those enzymes could be investigated at the same time, making the identification of truly differentiated ligands with desired mode of action (MoA) possible. In our previous series, DEL selection for enzymes like kinases and DNA binding proteins has already been reviewed, and therefore in this chapter we mainly focus on enzymes with multi-substrates and unique reaction mechanisms, as well as the ones that bind to specific undesired moieties.
DEL selection for enzymes with compulsory ordered reaction mechanism
While most activity-based screenings yield functional hits, the affinity-based DEL selection presents unique advantages by providing ligands with desired MoA and therefore higher likelihood of successful drug efficacies. This is particularly beneficial for those targets with multiple substrates. For example, Naa50, an enzyme that utilizes a compulsory ordered reaction pathway, in which acetyl-CoA binds first, revealing a binding pocket for the recognition by a second substrate. The detailed understanding of this reaction mechanism helped us design a delicate DEL selection plan, leading to the identification of hits with single digit nM potency (ACS Med Chem Lett. 2020 Apr 10;11(6):1175-1184, figure 1).
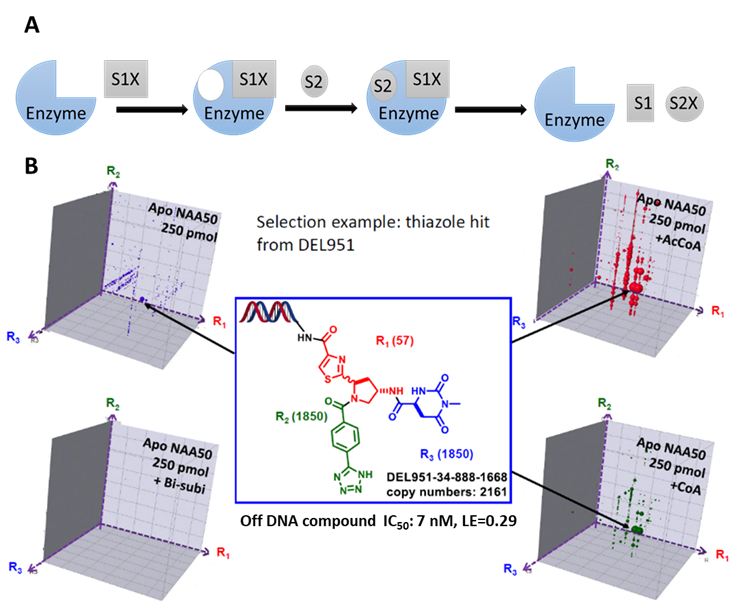
Figure 1, A) Compulsory ordered reaction pathway. B) Naa50 inhibitor identified from DEL selection with a unique binding mechanism.
DEL selection for enzymes with ping-pong reaction mechanism
Ping-pong mechanism is another typical enzymatic reaction pathway that proceeds in two sequential steps. During such a reaction, the enzyme first catalyzes the turnover of one substrate to form an enzyme-X intermediate complex, and then releases the first product; only after that the second substrate could bind to the enzyme and proceed to generate and release the second product. At HitGen, DEL selections are carefully designed to include the intermediate forms of interested targets in addition to the apo enzyme, which offers unique opportunities to investigate the target structures that represent distinct conformations. Therefore, we significantly increase the chance to identify hits with desired MoA (Figure 2).
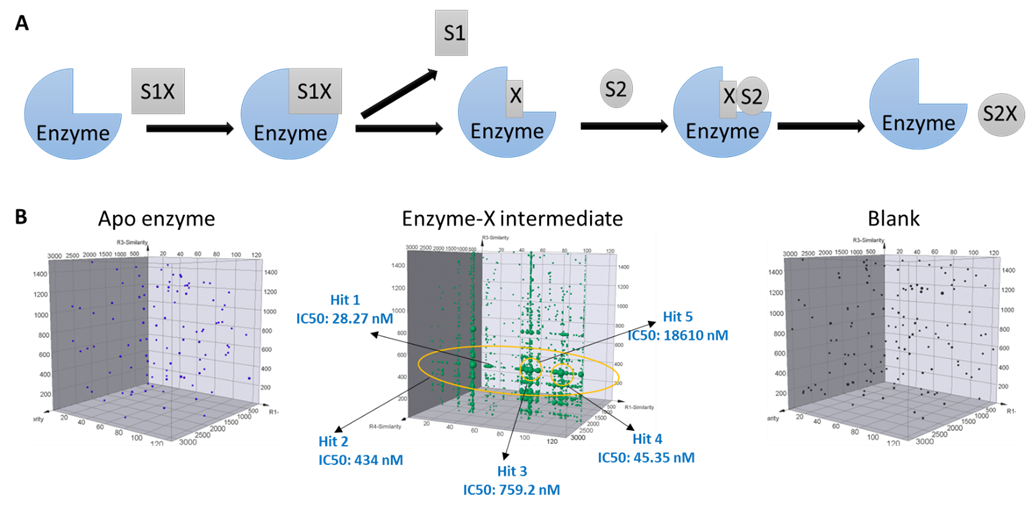
Figure 2, A) Ping-pong reaction pathway. B) DEL selection signal comparison for different forms of target, and multiple inhibitors were identified from DEL selection for an enzyme target with ping-pong mechanism.
DEL selection for enzymes with desired competition mechanism
Competitive, non-competitive, and uncompetitive inhibitions are major mechanisms of reversible inhibitors. A selection that includes a physiological substrate or a well-characterized ligand of the interested enzyme could help identify hits with different competition MoAs. On the other hand, some targets such as tyrosine phosphatases might have basic or acidic binding pockets or interaction moieties. Although it is relatively easy to identify binders for these targets, further optimization from these hits for pharmaceutically acceptable ligands is usually challenging due to their poor physical/chemical properties. Incorporating a catalytic site binder during DEL selection could help rule out identification of more active site binders, but to allow the identification of binders to potential allosteric sites. With such a design, a DEL selection offers opportunities to identify hits with more desired pharmaceutical properties. Figure 3 illustrates an example that utilizes active site blocker to identify ligands binding to allosteric sites.
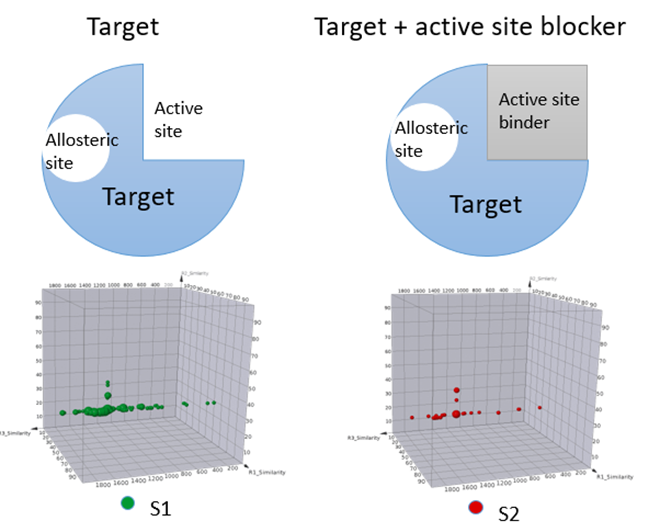
Figure 3, Schematic of including active site binders to differentiate allosteric site binders.
Enzymes are the primary effectors in physiological conditions and account for nearly half of drug targets, and therefore identification of ligands targeting enzymes provide opportunities to cure different diseases. At HitGen, we offer DEL selections that help your exploration of enzyme targets with quick turn-around time, low cost and unique advantages:
l Investigation of enzymes at specific conformations or distinct structures.
l Convenient exploitation of enzyme ligands at orthosteric and allosteric sites.
l Quick inclusion of counter targets at low expenses.
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information