One-stop Technology Platform to Accelerate Drug Discovery and Development
Protein degraders (most popularly known as PROTAC or Molecular Glue) utilize an event-driven process, recruit protein of interest (POI, or targeted protein) and E3 ligase directly or indirectly, induce poly-ubiquitination of POI and subsequent proteasome-mediated degradation. Unlike protein inhibitors, protein degraders do not require inhibitory activity to the protein, therefore, this modality of therapeutics applies to broader target classes including undruggable proteins, scaffold proteins, mutant proteins and etc.
PROTAC
Proteolysis Targeting Chimera (PROTAC) is a bivalent molecule consisting of a POI binder, an E3 ligase binder, and a linker tethering the two binders. Transferring an inhibitor and known E3 ligase binders (most commonly used ones are CRBN and VHL binders) with suitable linkers has been very popular in the past couple of years and yielded several clinical PROTAC programs (Figure 1). The efficacy against mutations, lower dosage and long-lasting effect highlights the PROTAC advantages over direct inhibitors.
Molecular Glue
Molecular glue (MG) is a monovalent molecule that brings two proteins in proximity as if they are glued together. This mechanism of action has been reported in different contexts. In protein degradation case, it induces POI:MG:E3 complex formation with the following several possible events: 1) MG significantly enhances POI:E3 interaction; 2) MG binds to POI, followed by recognition of E3; 3) MG binds to E3 ligase to allow the recognition of neosubstrate. Only proper binding pose(s) between POI and E3 allow ubiquitination and proteasome degradation. Molecular glue is a desired protein degrader, but it’s much more challenging for discovery or design, which is evidenced with less targets and programs in the clinical.
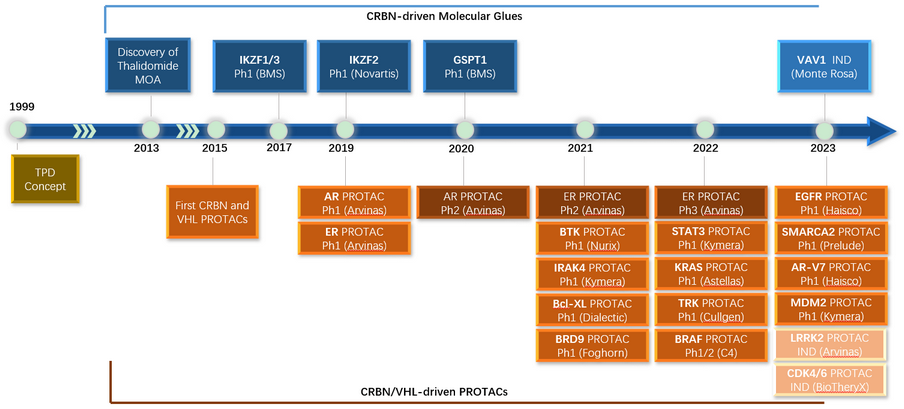
Figure 1. Protein degrader discovery history
DEL for PROTAC based selection
DNA Encoded Library (DEL) is a very powerful technology for protein binder discovery because of its ability to access vast number of chemical compounds (HitGen full DEL contains more than one trillion compounds) with efficient protein usage, rapid turn around and low cost. Multiple selection experiments and deep data analysis ensure that the binders identified are selective to the POI. Also, due to the nature of DEL compounds, the binders discovered by DEL screening have a natural attachment site for building linkers of a PROTAC molecule. We have recently developed a method to optimize POI:degrader:E3 complex formation with POI, E3 dual protein selection, the PROTAC DEL and corresponding selection are published in ACS Chem. Biol. 2023, 18, 25-33 (ACS Editors’ Choice, figure 2).
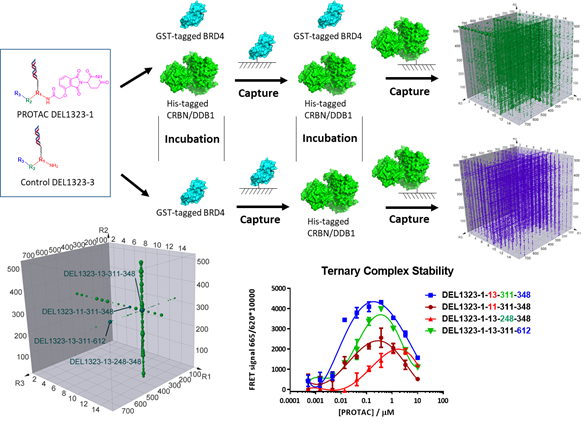
Figure 2, DEL technology for ternary complex optimization
PROTAC and MG development from DEL
PROTAC discovery and development are now beyond oncology disease area hence tissue/cell restricted E3 ligase expression has become a very useful way for differentiation. As mentioned above, DEL technology is very effective for binder discovery, even in the situation where protein function is not well understood like in the case of most of E3s. Several dozens of purified E3s have been expressed, purified and screened against HitGen DELs, yielding many specific binders especially for those E3s without any binders reported. These binders are gradually approved for their PROTAC functions for important proteins of interest. During the exploration of PROTAC degradation, some of these E3 binders have been found with potential molecular glue function (More details will be given upon request).
PROTAC development from DEL derived CRBN binders
Emerging from the reported CRBN binders, novel CRBN binders discovered from DEL selection showed higher binding affinity, more potent degradation with its PROTACs against many popular targets, and no IMiD (immunomodulatory drug) function (Figure 3).
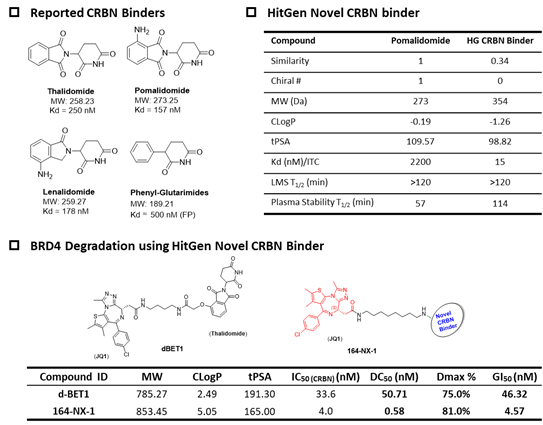
Figure 3, Novel DEL-derived CRBN PROTACs for potent POI degradation
PROTAC development from DEL derived BIRC7 binders
BIRC7 (specifically expressed in cancer cells) ligand is the very first molecule identified from HitGen 1T DEL screening and its PROTAC shows selective degradation in BIRC7 high expression cell line (Figure 4).
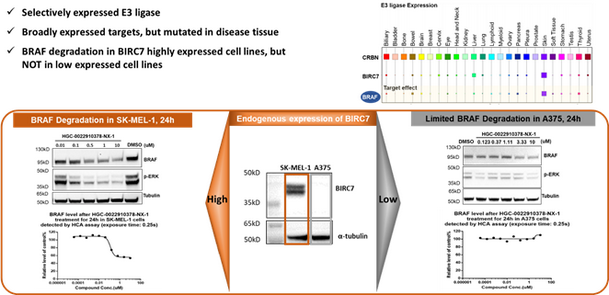
Figure 4, BIRC7 based PROTAC affords selective target degradation
PROTAC development from DEL derived TRIM21 binders
TRIM21 (broad expression but different from CRBN) binders discovered from HitGen DEL screening have been developed into PROTACs and found with very potent BRD4 degradation (pM DC50, 8000X more potent than CRBN based PROTAC), differentiated kinase degradation comparing to CRBN based PROTACs, more importantly, TRIM21 ligands have potential Molecular Glue function (Figure 5).
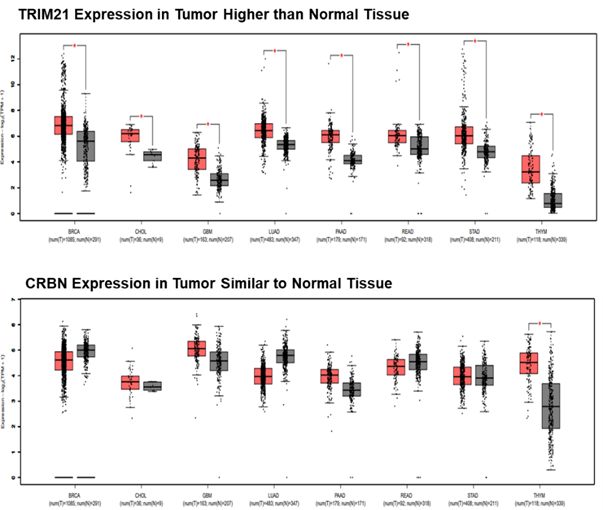
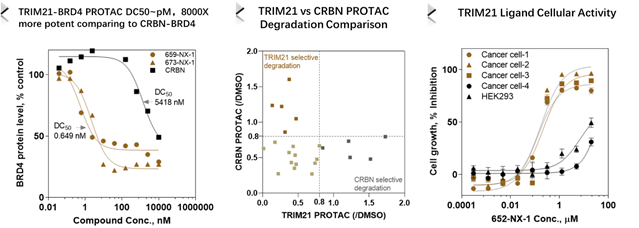
Figure 5, TRIM21 based PROTAC offers more potent and different degradation profile
DEL selection for protein degraders at HitGen
Along with our trillion size DEL and PROTAC DEL, HitGen provides customized selection strategies to ensure a successful screen for protein degrader discovery:
● HitGen trillion DEL offers large chemical space for binder identification, independent of protein activity.
● PROTAC DEL provides ternary complex based selection.
● Multiple E3 ligases incorporated in one selection give high throughput and selectivity.
● Linker position is known for PROTAC development.
● Track record of successful selection cases with novel ligands identified.
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information